Reshaping and Plotting data
Overview
Teaching: min
Exercises: minQuestions
Objectives
Learning Objectives
- Reshape a tibble
- Start with ggplot2
Our biological question: can we see the behaviour of our favourite genes?
- Plot the individual trajectories of favourites: NOP16, NOP56, ACT1
Reshape/tidy the data
gather(mRNA_data,key=Vol,value="log2_ratio",-Name,-Gene)
# A tibble: 53,704 x 4
Name Gene Vol log2_ratio
<chr> <chr> <chr> <dbl>
1 Q0010 <NA> 40 fL 0.145
2 Q0017 <NA> 40 fL -1.32
3 Q0032 <NA> 40 fL NA
4 Q0045 COX1 40 fL 0.252
5 Q0050 AI1 40 fL -0.784
6 Q0055 AI2 40 fL -0.369
7 Q0060 AI3 40 fL -0.143
8 Q0065 AI4 40 fL -0.377
9 Q0070 AI5_ALPHA 40 fL -0.439
10 Q0075 AI5_BETA 40 fL -0.450
# ... with 53,694 more rows
mRNA_data_gathered <-
gather(mRNA_data,key=Vol,value="log2_ratio",-Name,-Gene) %>%
separate(Vol,into="Vol_fL") %>%
mutate(Vol_fL = as.numeric(Vol_fL))
Warning: Expected 1 pieces. Additional pieces discarded in 53704 rows [1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, ...].
mRNA_data_gathered
# A tibble: 53,704 x 4
Name Gene Vol_fL log2_ratio
<chr> <chr> <dbl> <dbl>
1 Q0010 <NA> 40 0.145
2 Q0017 <NA> 40 -1.32
3 Q0032 <NA> 40 NA
4 Q0045 COX1 40 0.252
5 Q0050 AI1 40 -0.784
6 Q0055 AI2 40 -0.369
7 Q0060 AI3 40 -0.143
8 Q0065 AI4 40 -0.377
9 Q0070 AI5_ALPHA 40 -0.439
10 Q0075 AI5_BETA 40 -0.450
# ... with 53,694 more rows
mRNA_data_NOP16 <- filter(mRNA_data_gathered, Gene=="NOP16")
mRNA_data_NOP16
# A tibble: 8 x 4
Name Gene Vol_fL log2_ratio
<chr> <chr> <dbl> <dbl>
1 YER002W NOP16 40 0.687
2 YER002W NOP16 45 0.706
3 YER002W NOP16 50 0.203
4 YER002W NOP16 55 -0.0884
5 YER002W NOP16 60 -0.698
6 YER002W NOP16 65 -0.708
7 YER002W NOP16 70 -1.05
8 YER002W NOP16 75 -0.0681
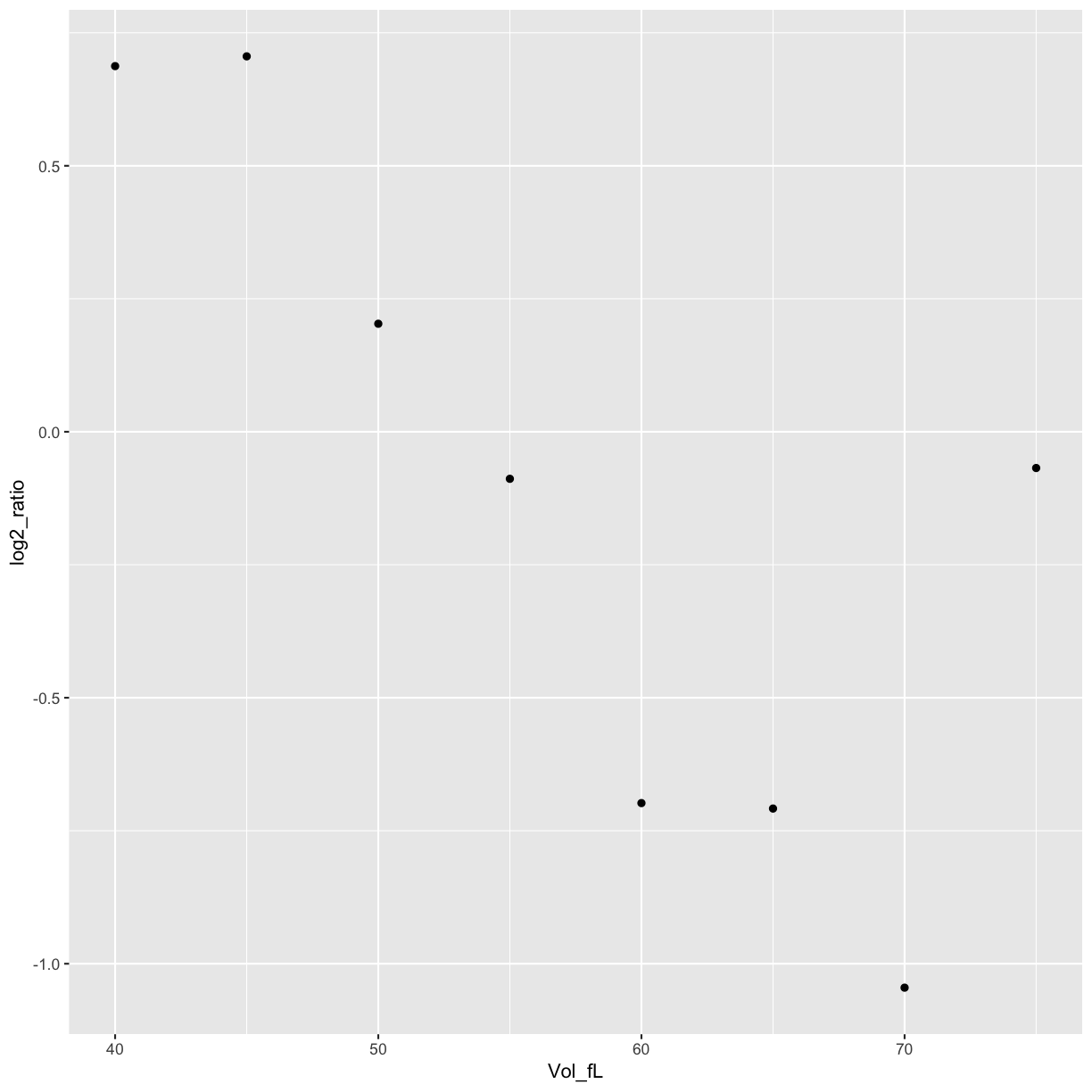
Plot NOP16 data
ggplot(data=mRNA_data_NOP16, aes(x=Vol_fL,y=log2_ratio)) +
geom_point()
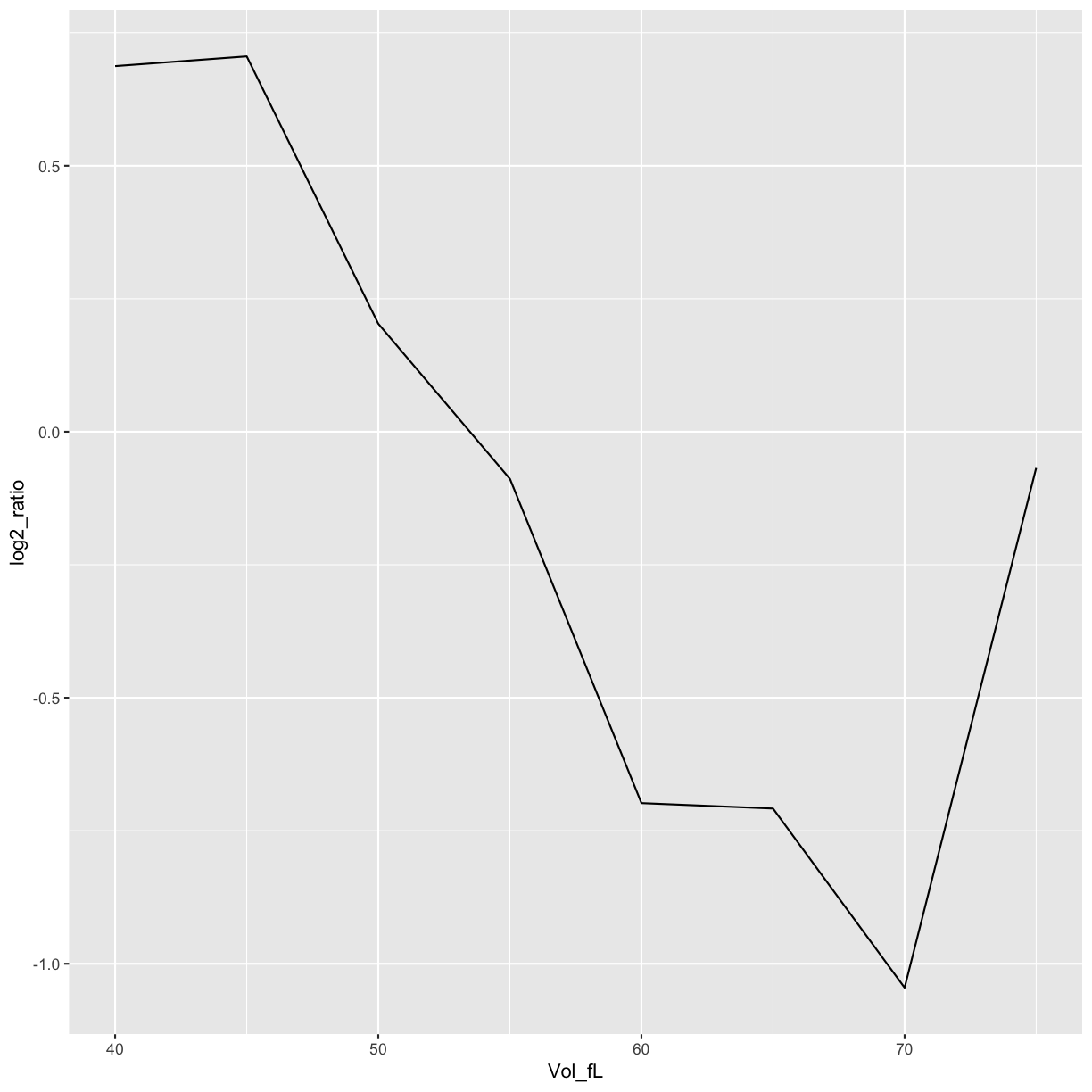
ggplot(data=mRNA_data_NOP16, aes(x=Vol_fL,y=log2_ratio)) +
geom_line()
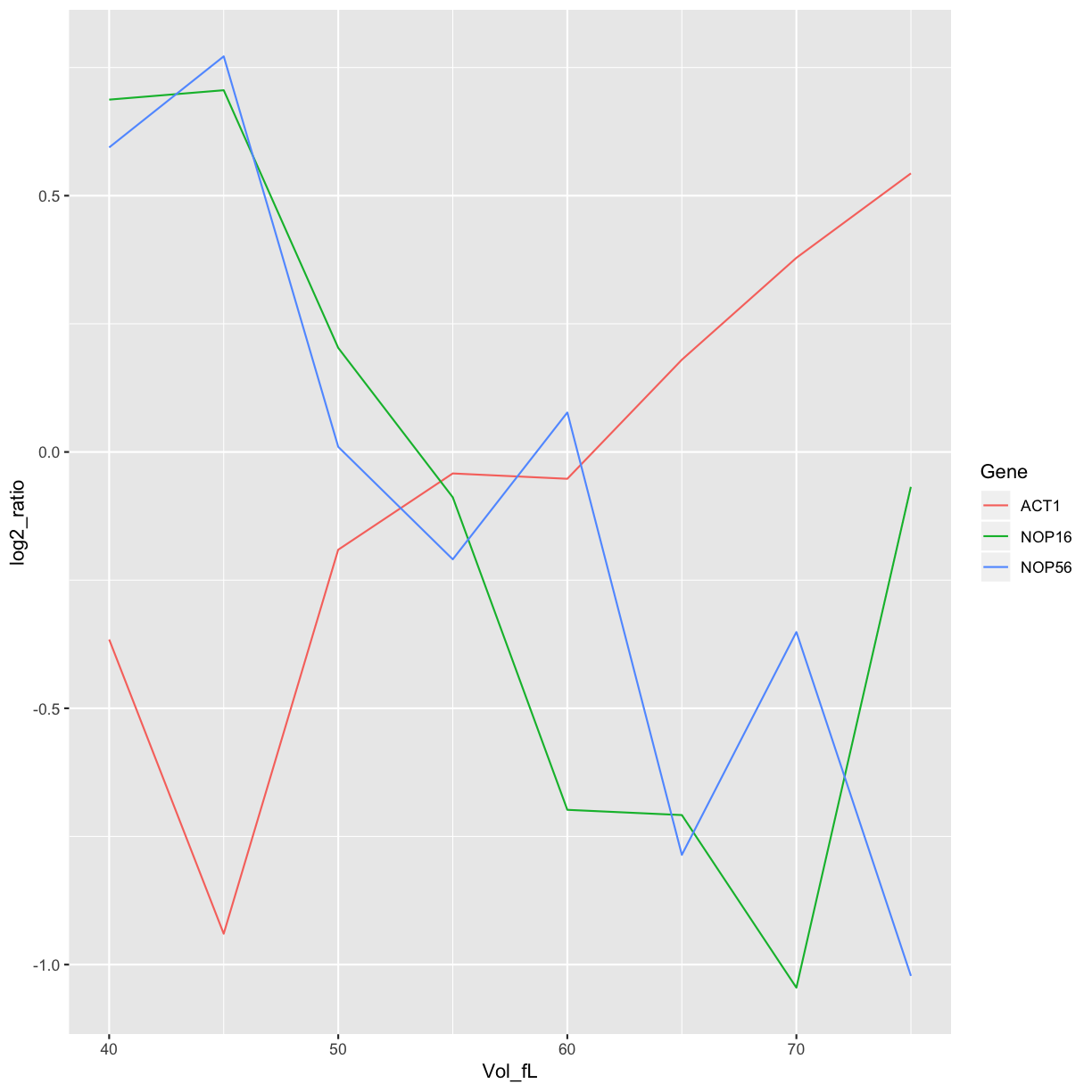
Plot all the favourite genes data
mRNA_data_3genes <- filter(mRNA_data_gathered, Gene %in% c("ACT1","NOP16","NOP56"))
ggplot(data=mRNA_data_3genes, aes(x=Vol_fL,y=log2_ratio,colour=Gene)) +
geom_line()
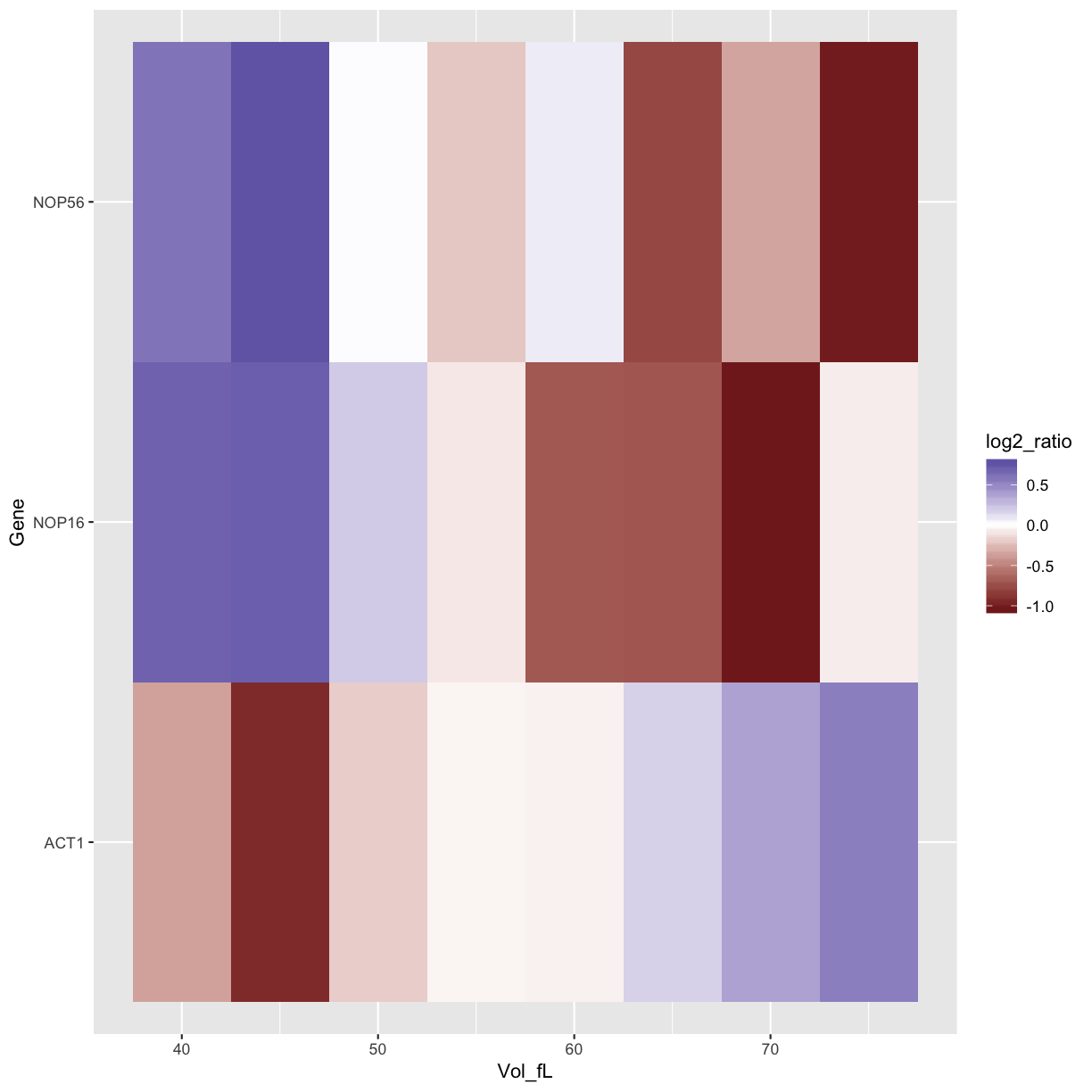
ggplot(data=mRNA_data_3genes, aes(x=Vol_fL,y=Gene,fill=log2_ratio)) +
geom_tile()
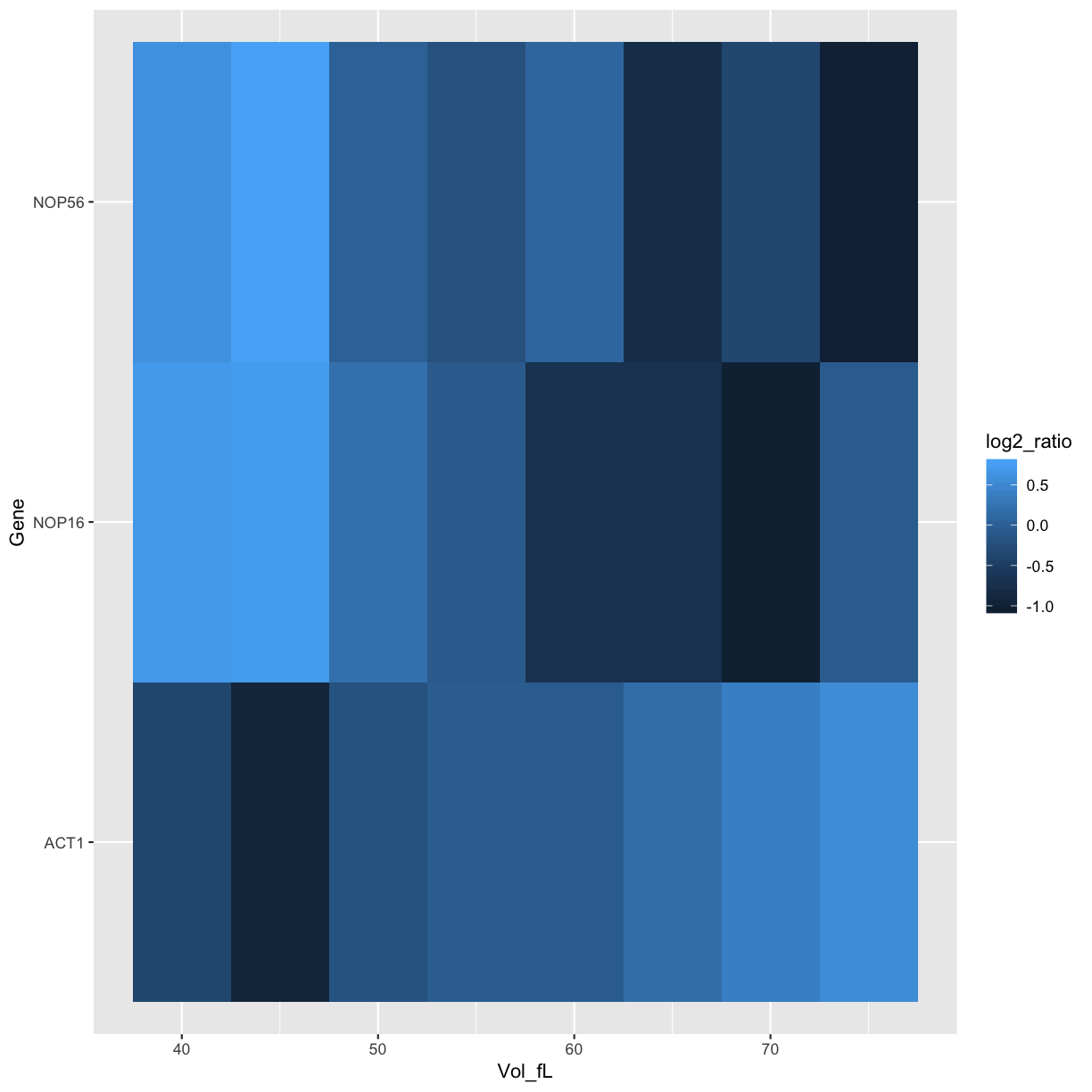
ggplot(data=mRNA_data_3genes, aes(x=Vol_fL,y=Gene,fill=log2_ratio)) +
geom_tile() +
scale_fill_gradient2()
How to make the plot look nice
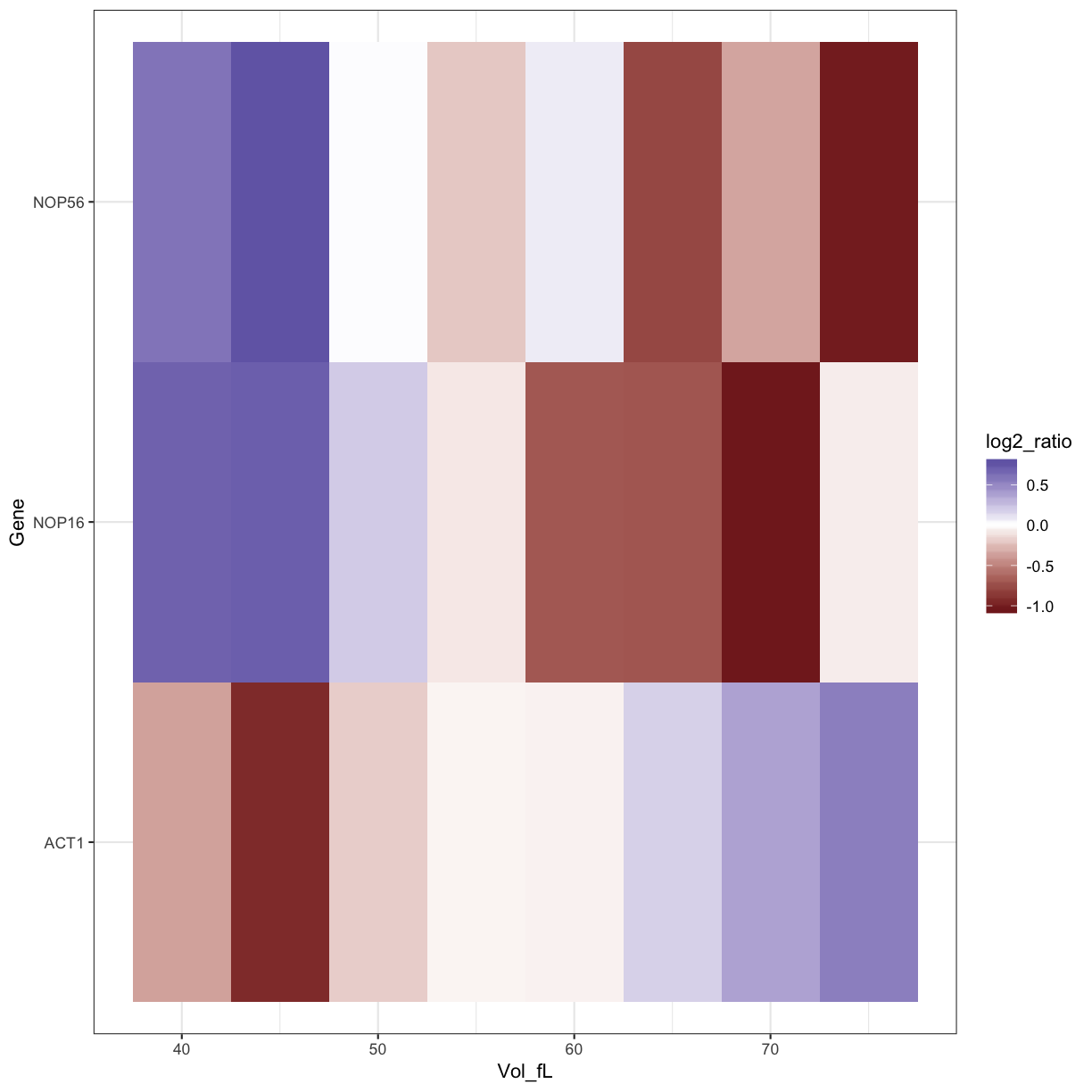
ggplot(data=mRNA_data_3genes, aes(x=Vol_fL,y=Gene,fill=log2_ratio)) +
geom_tile() +
scale_fill_gradient2() +
theme_bw()
Challenge:
- Plot your other favourite genes. NOP6?
- Can you change the line colours to something nicer?
Solution
mRNA_data_4genes <- filter(mRNA_data_gathered, Gene %in% c("ACT1","NOP16","NOP56","NOP6")) # ?scale_colour_brewer
Key Points